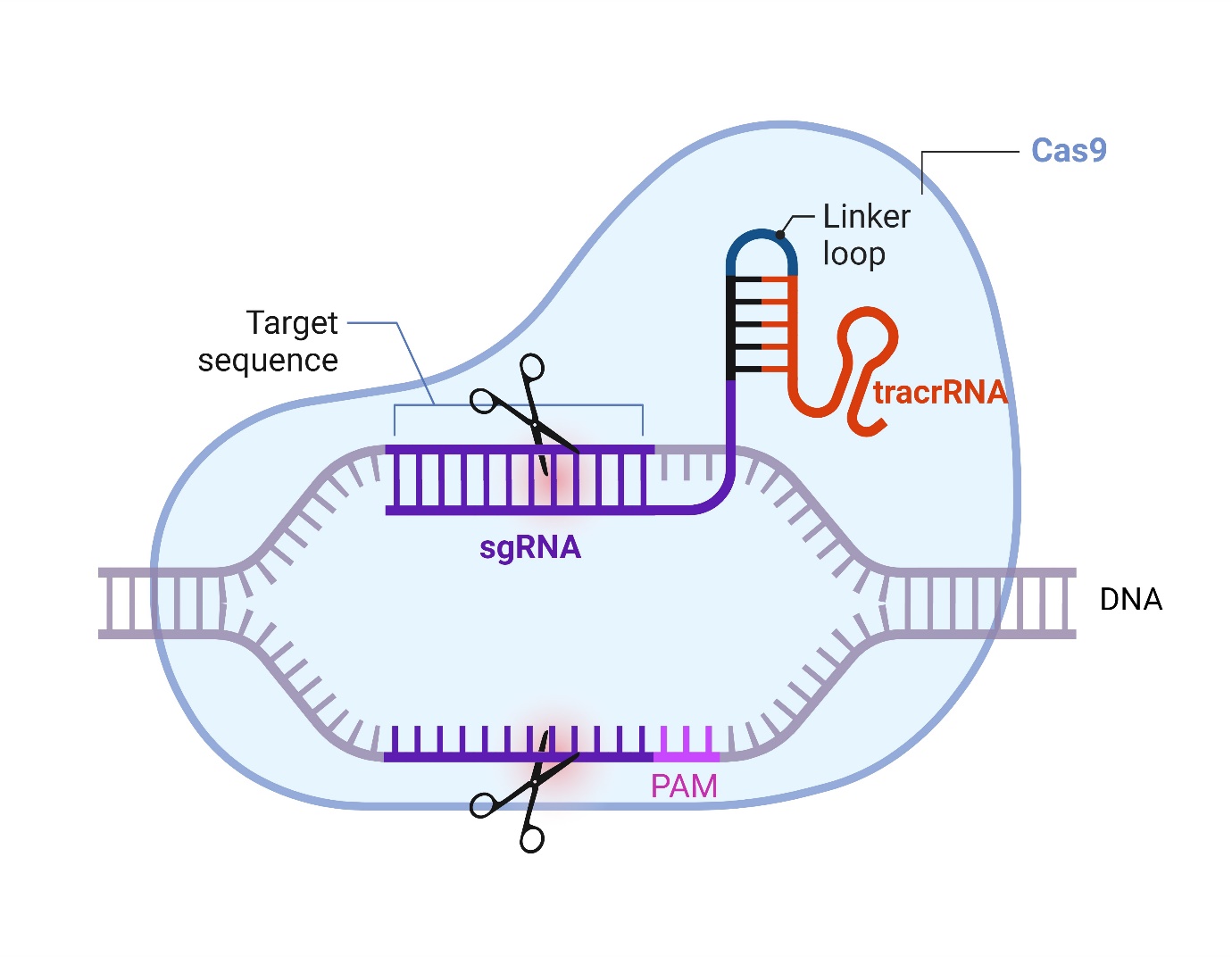
The CRISPR-Cas9 system has revolutionized the field of molecular biology and disease modelling. It is a method of editing the genetic code that is derived from processes originally observed in the bacterial immune system. It comprises a CRISPR-associated protein 9 (Cas9) endonuclease, a trans-activating CRISPR RNA (tracrRNA), and a single-guide RNA (sgRNA) that recognises a target sequence downstream of a protospacer adjacent motif (PAM) sequence (5’-NGG-3’).
Previous methods employed to edit the genome such as zinc-finger nucleases or transcription activator-like effector nucleases are time consuming, laborious and can be associated with off-target effects. The CRISPR-Cas9 system relies on DNA-RNA interactions to regulate target sequence recognition, making it easy to locate, remove, add, or alter sections of genetic code with minimal off-target effects.
CRISPR also benefits from a simplified cloning approach that employs a small recognition sequence of the sgRNA (20 base pairs) relative to the much larger approaches used in traditional homologous repair design.
